Hemp breeding
Brian J. Knaus
2024, November 08
Cannabis sativa L. has been used almost as long as there has been civilization (Cannabis at Wikipedia). The plant fiber has been used as a textile to make cloth, it’s seeds have been used as food for humans and animals, and it’s essential oils have been used as medicine. The 2018 U.S. farm bill decriminalized industrial hemp and has defined it as plants containing less than 0.3% \(\Delta\)(9)-tetrahydrocannabinolic acid. A goal of the lab is to use genomics to better understand agronomically important traits and how we can make informed decisions to breed the best crops possible.
Hemp genomics.
The hemp genome is thought to be aroun 800-900 Mbp (haploid size) and consist of nine pairs of autosomes and a pair of sex chromosomes. The Kew Plant C-values database reports Cannabis sativa as having a 1C (pg) of 0.84, or 821.52 Mbp (Wikipedia C-value conversion). Sequencing efforts have resulted in over ten genomic assemblies of various qualities assembled with a variety of technologies (Cannabis at NCBI). A recent attempt (Grassa et al. 2021) resulted in an assembly of all ten chromosomes (as well as unplaced contigs). Building on these genomic resources we hope to learn to better understand how these plants produce essential oils as well as other phytochemicals.
nuc <- structure(list(Id = c("NC_044371.1", "NC_044375.1", "NC_044372.1",
"NC_044373.1", "NC_044374.1", "NC_044377.1", "NC_044378.1", "NC_044379.1",
"NC_044376.1", "NC_044370.1"), Length = c(101209240, 96346938,
94670641, 91913879, 88181582, 79335105, 71238074, 64622176, 61561104,
104987320), a = c(12762692, 15648113, 15834625, 15706376, 16864131,
15448470, 12544227, 9979448, 10305137, 16150029), A = c(11748277,
9880538, 9810980, 10180981, 10000065, 9320665, 7534637, 8191831,
7518942, 12415232), c = c(4717714, 6495082, 6494817, 6294060,
7007138, 6429275, 5306040, 3917350, 4089354, 6429511), C = c(7349355,
6457816, 6342764, 6498433, 6511470, 6046421, 4942448, 5190552,
4771093, 8027053), g = c(4737281, 6501151, 6541912, 6321483,
6996441, 6460883, 5307013, 3917059, 4120057, 6440113), G = c(7366441,
6462266, 6351808, 6511484, 6528963, 6025024, 4953814, 5184073,
4778222, 8042296), t = c(12761506, 15640696, 15877710, 15713009,
16816817, 15442514, 12513884, 9980581, 10312600, 16153769), T = c(11753649,
9888880, 9820939, 10176745, 9975902, 9337925, 7535448, 8161118,
7493785, 12429366), n = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0), N = c(28012325,
19372396, 17595086, 14511308, 7480655, 4823928, 10600563, 10100164,
8171914, 18899951)), row.names = c(NA, -10L), class = c("tbl_df",
"tbl", "data.frame"))
# nucchrmw <- 0.05
prbw <- 0.2
plot(1:nrow(nuc), nuc$Length/1e6, type = "n", xlab = "", ylab = "",
ylim = c(0, max(nuc$Length)/1e6), xaxt = "n")
axis(side = 1, at = 1:10, labels = c(1:9, "X"))
title(xlab = "Chromosome")
title(ylab = "Position (Mbp)")
rect(xleft = 1:nrow(nuc) - chrmw, ybottom = rep(1, times = nrow(nuc)),
xright = 1:nrow(nuc) + chrmw, ytop = nuc$Length/1e6,
col = "#C0C0C0",
border = "#808080")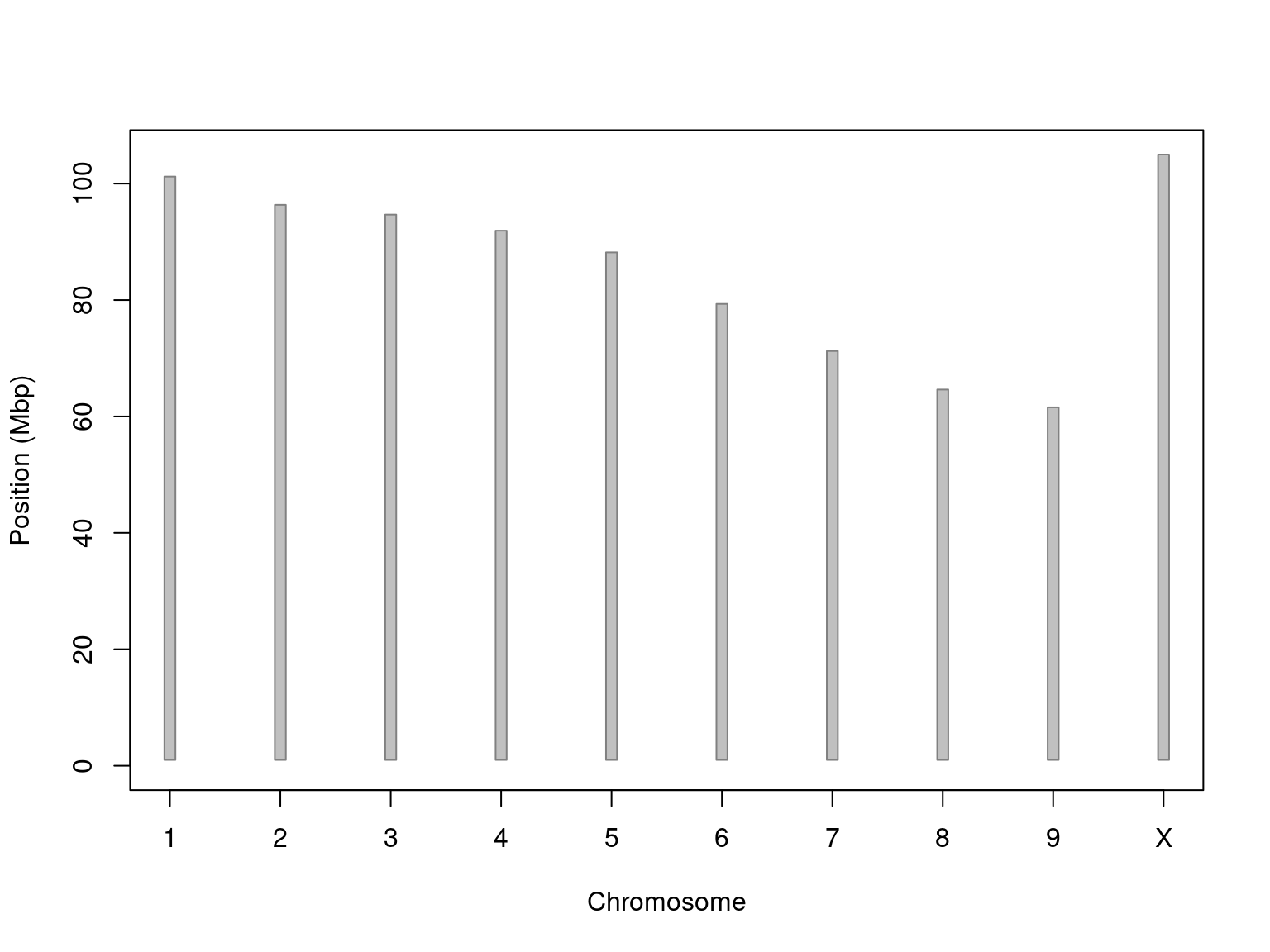
Figure 1. Idiogram of the nine CBDRx autosomal chromosomes and a sex chromosome (CBDRx was genetically female so it was homozygous for the X chromosome).
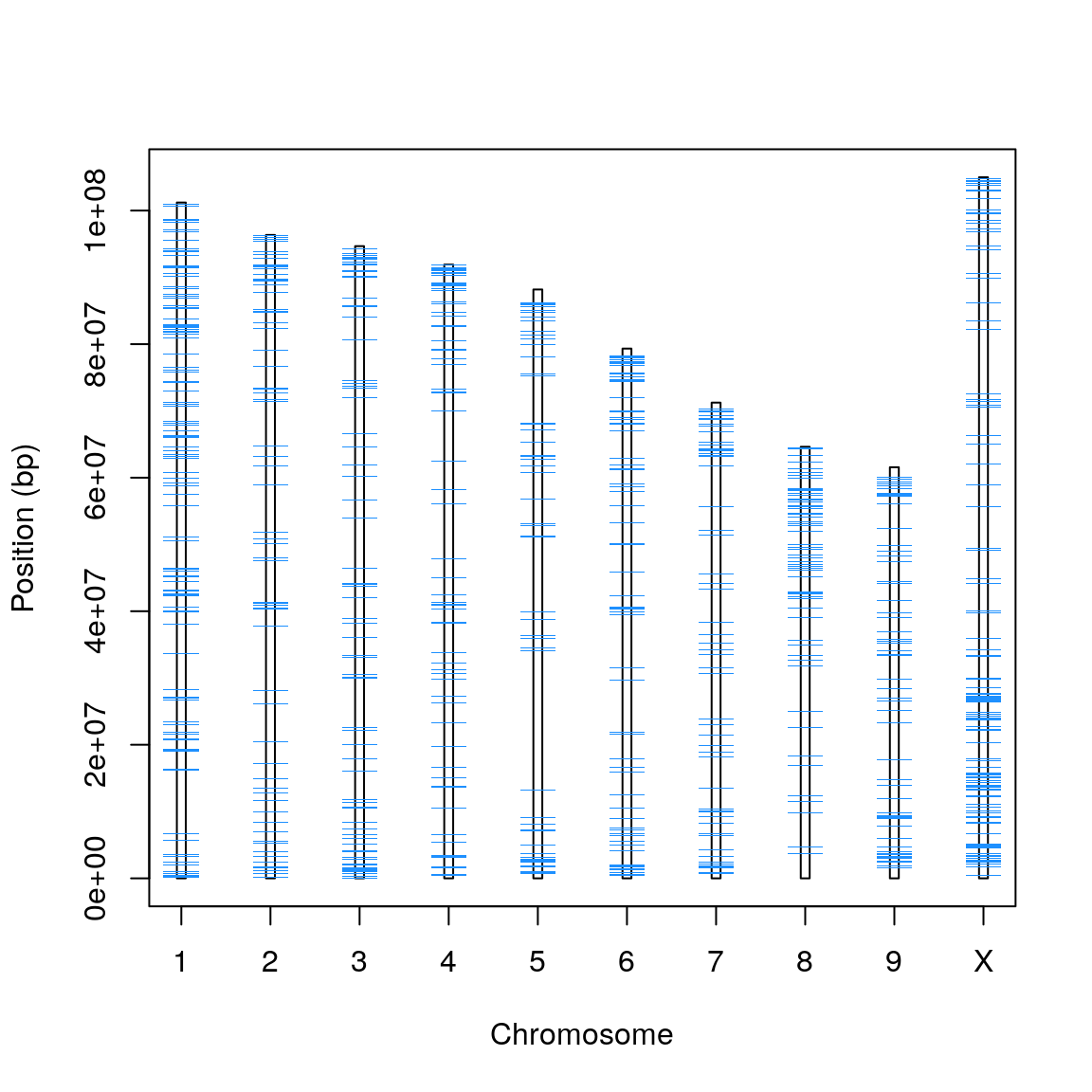
Figure X. The chromosomes of CBDRx with mRNAs depicted in light blue.
Internet Links
International Cannabis Genomics
Research Consortium
The Cannabis Genome
Browser, the University of Toronto
Cannabis at
Wikipedia
Cannabis GDB
Kannapedia
Hemp pests
at Pacific Northwest Pest Management Handbooks
Comparative Genomics CoGe
Cannabis
Oregon Department of Agriculture hemp
program
Industry links
Strain information:
CBD
Logistics (Switzerland) - Certified seeds
Leafly (WA, USA) -
Strain lists
Dewey Scientific (WA,
USA)